Protein-Ligand Interactions
| Logo | Name | Description | Authors | Additional sponsors |
|---|---|---|---|---|
 |
@TOME V2 | Bioinformatics Tools Server: pipeline for comparative modeling of protein-ligand complexes | Pons JL and Labesse G, NAR (2009); doi : 10.1093/nar/gkp368 | ACI-IMPBio |
| EDMon v3 | EDMON v3 | Endocrine Disruptor MONitoring Computational tool to study interactions between nuclear hormone receptors and endocrine-disrupting chemicals |
Delfosse V et al, PNAS, 109, 14930-5 (2012) - Doi : 10.1073/pnas.1203574109 | ANR BISCOT |
 |
CoSiAn | Combinatorial Similarity Analysis Tool for similarity analysis between a reference ligand and a bank of ligands to screen, using a combination of 2D and 3D similarity tools |
Reys V, Moreau V, Pons J-L, Labesse G | |
 |
CBE database Visualizer | Tool to visualize Protein Chain/Ligand contacts in PDB file (Ligand can be small molecule, sugar, nucleic acid or peptide) | Pons J-L and Labesse G | |
 |
KinDOCK | Ligand Transposition Server: comparative docking of ligands into the ATP-binding site of a protein kinase (target) | Martin L, Catherinot V, Labesse G, NAR Web Server, issue:34 W325-W329 (2006) | La Ligue contre le cancer |
 |
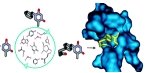
SimDOCK | Ligand Transposition Server: rapid selection of ligands fitting the active site of the submitted protein by superposition of its three-dimensional structure with those of known complexes of protein/ligands of the family | Martin L, Catherinot V and Labesse G | La Ligue contre le cancer |
 |
LEA3D | Computer-Aided Ligand Design for Structure-Based Drug Design | Douguet D, Munier-Lehmann H, Labesse G and Pochet S J, J Med Chem, 48(7), 2457-68 (2005) | |
 |
TSA-CRAFT | Thermal Shift Analysis | Lee PH, Huang XX, Teh BT, Ng LM. SLAS Discov. 2019;24(5):606-612. Webserver Implementation by V.Reys |