NMR Tools
| Logo | Name | Description | Authors | Additional sponsors |
|---|---|---|---|---|
 |
CCSP | Consensus for Chemical Shift Prediction (based on the following softwares: Shifty v1.4, Orb v1.2, Shifts v4.1.1, Proshifts and Shiftx) | Pons JL and Delsuc MA | ACI-IMPBio |
 |
CINDY | Graphical NMR assignment software | Padilla A | |
 |
CRAACK | Consensus Rules for Amino-Acid Characterisation using a Kernel method (SVM); the spin system typing consensus is based on the following softwares: Rescue, RescueN, Rescue2, Platon, and one other using SVM algorithm | Benod C, Delsuc MA, Pons JL, J Chem Inf Model, 46 (3), 1517-1522 (2006) | ACI-IMPBio |
 |
CROWD | Noe intensity simulation software | Malliavin T, J Biomol NMR, 5, 193-201 (1995) | |
 |
DExt | Tool for distance extraction from PDB file | Martin L, Pons JL, Delsuc MA | IMPG |
| DYNAMOF | DYNAMOF | MATLAB program devoted to the analysis of relaxation data, possibly obtained at different magnetic field values | Barthe P and Roumestand C | |
 |
GIFA | Computer program designed for the processing, visualization and analyses of 1D, 2D, and 3D NMR data-sets | * Pons JL, Malliavin T, Delsuc MA, J Biomol NMR, 8, 445-452 (1996) * Malliavin T, Pons JL, Delsuc MA, Bioinformatics, 14, 624-631 (1998) |
NMRtec |
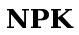 |
NPK | NMR Processing Kernel, a python based NMR processing program: replacement for the Gifa project, still in beta though; fully pipeline-able | Delsuc M-A, Catherinot V and Tramesel D | NMRtec |
 |
Rescue2 | Program for amino acid type assignment based on a probabilistic model of NMR chemical shifts distributions | Marin A, Malliavin TE, Nicolas P, Delsuc MA, J Bio NMR. 30, 47-60 (2003) | ACI-IMPBio |
| RESCUE (and RescueN) |
NMR residu typing assignment aid software based on Neural Networks | * Pons JL, Delsuc M-A, J Bio NMR, 15(0), 15-26 (1999) * Auguin D, Catherinot V, Malliavin TE, Pons JL and Delsuc MA, Spectroscopy, 17(0), 559-568 (2003) |
IMPG |