ViTO 2
A tool for homology modelling and protein family
study
Download and installation
Running the application locally
Start ViTO2 online
Screenshots
User guide
Introduction
ViTO2 is a rewrite and a merge of ViTO and
TiTO tools.
It is available as a GUI tool and a command line tool.
Main handled file formats are PIR
for alignments and PDB
for protein structures.
As a graphical tool, it interactively allows to modify sequence
alignments (some sequences having an associated experimental
structure, used as template) while displaying the generated
protein model and evaluating the model quality through potential
of mean force (PKB) score.
As a command line tool, it can generate a PDB 3D protein model
from existing structure and a given PIR alignment, also computing
PKB score on the model.
Vito2 is also integrated as protein alignment refining tool in
the @tome platform.
Installation
Download GUI version, console version, and installation documentation (french version).
Running
These steps apply after downloading and installation on your own
computer. To start Vito2 directly from this page, see this section.
In any case, Java 7 or higher is required.
GUI version
From desktop, double click on vito2_gui.jar in the installation
directory.
Alternatively, from command line :
- either cd to the installation directory and run :
- ./vito2-gui <alignment input file> (Linux/MacOsX)
- .\vito2-gui <alignment input file> (Windows)
- or, if PATH variable has been configured, you can run the
application from anywhere with :
- vito2-gui <alignment input file> (Linux/MacOsX)
- vito2-gui <alignment input file> (Windows)
Command line version
From command line :
- either cd to the installation directory and run :
- ./vito2-console <arguments> (Linux/MacOsX)
- .\vito2-console <arguments> (Windows)
- or, if PATH variable has been configured, you can run the
application from anywhere with :
- vito2-console <arguments> (Linux/MacOsX)
- vito2-console <arguments> (Windows)
Command syntax is (you can get it by simply running vito2-console
with no argument) :
- vito2-console <PIR alignment file> [-t <arg>] [-s
<arg>] [-w <arg>] [--p2d <arg>] [--warn][-d]
- -t,--target <arg> : name of sequence to model
- -s,--support <arg> : template structure name
- -w,--scwrl <arg> : generate SCWRL files (.pdb &
.scw) with given base file name. If no argument is given, base
will be <alignment name>-<target name>-<query
name>
- --p2d <arg> : read '2D structure prediction from 1D'
file and output matching score with prediction from 3D
structure
- --warn : enable warning messages
- -d,--debug : debug log messages
Web start
Click here to start ViTO2 now.
Java security settings may prevent you from starting this way. To
solve this :
- Java 7 : open the Java configuration tool (ControlPanel),
click on the "Security" tab. Check the "Enable java content in
the browser" toggle, and select the lowest level (Medium).
Apply/OK and reattempt.
- Java 8: proceed as for Java 7. After selecting the security
level, click on "Edit site list", then "Add". In the "Location"
area, type : https://bioserv.cbs.cnrs.fr and hit Enter key
or click "Add". Confirm (click "Continue"), OK, and reattempt.
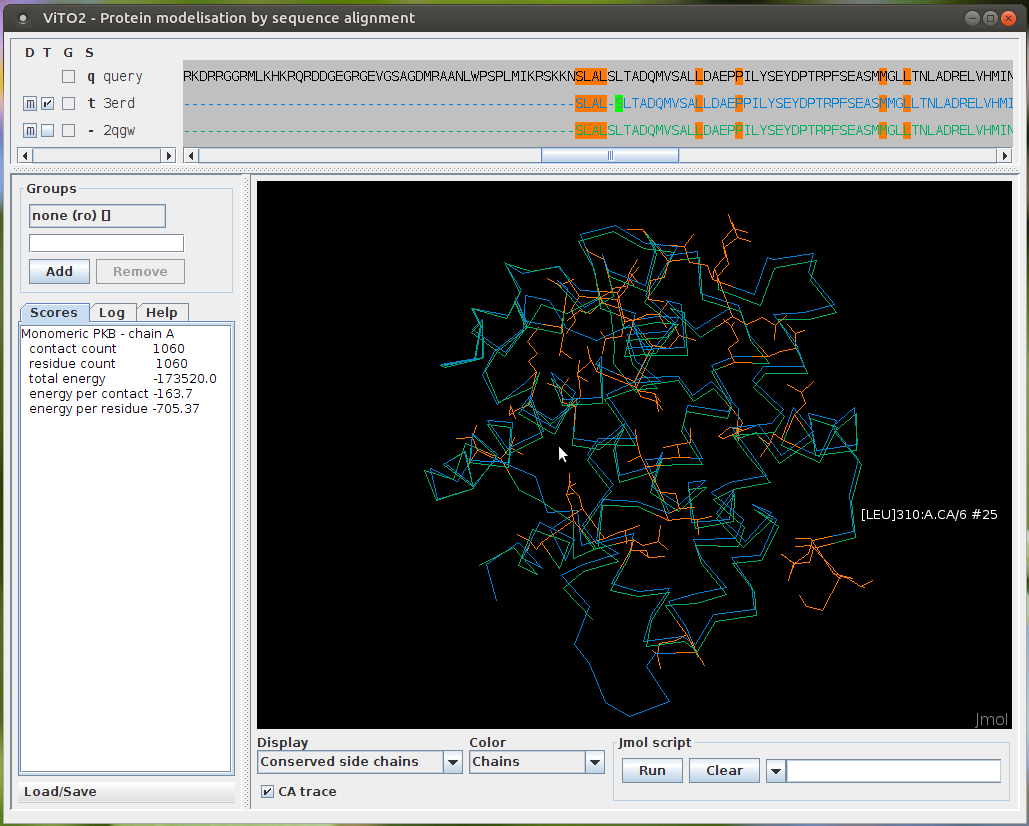
Screenshots
GUI main window
Console output
Example session of the command line version :