easYPipe ‘reindex’¶
This optional step is useful when several mtz should be in higher symmetry space group.
The program try to reindex according to the space group of the reference mtz.
Example: P422 can be re-indexed to P41212.
Usage¶
easypipe.py data reindex [-h] ref_mtz
arguments |
description |
|---|---|
-h, –help |
show this help message and exit |
sg_ref |
space group of reference for reindexing (name or number) |
-s, –simulate |
only simulate, generate a csv file listing for each process if mtz file will be reindexed or not. Give the possibility to modify the csv file to choose not to launch some reindexation, before launching again without simulation mode. |
Example:
$ easypipe.py PROCESSED_DATA reindex P41212
equivalent to:
$ easypipe.py PROCESSED_DATA reindex 92
What does it do ?¶
try to reindex mtz file with reflection_file_converter [1] if space group is different from reference space group


launch xtriage [2] for each successfully reindexed mtz to get resolution, completeness, space group and cell parameters
write a new ‘mtz_to_treat_ALL_reindexed.csv’ in ‘/easypipe/1b_reindex…’ folder, with reindexed mtz files information

becomes:

where P422, P4212 processed data have been successfully reindexed to P41212 space group.
Reindex simulation mode¶
Simulation mode allows to generate the csv file “reindex_mtz_<sg_ref>.csv” listing processes to be reindexed, but whithout launching reindexation. Then, you can modify the ‘to treat’ column to turn ‘yes’ to ‘no’ for some processes you don’t want to reindex .
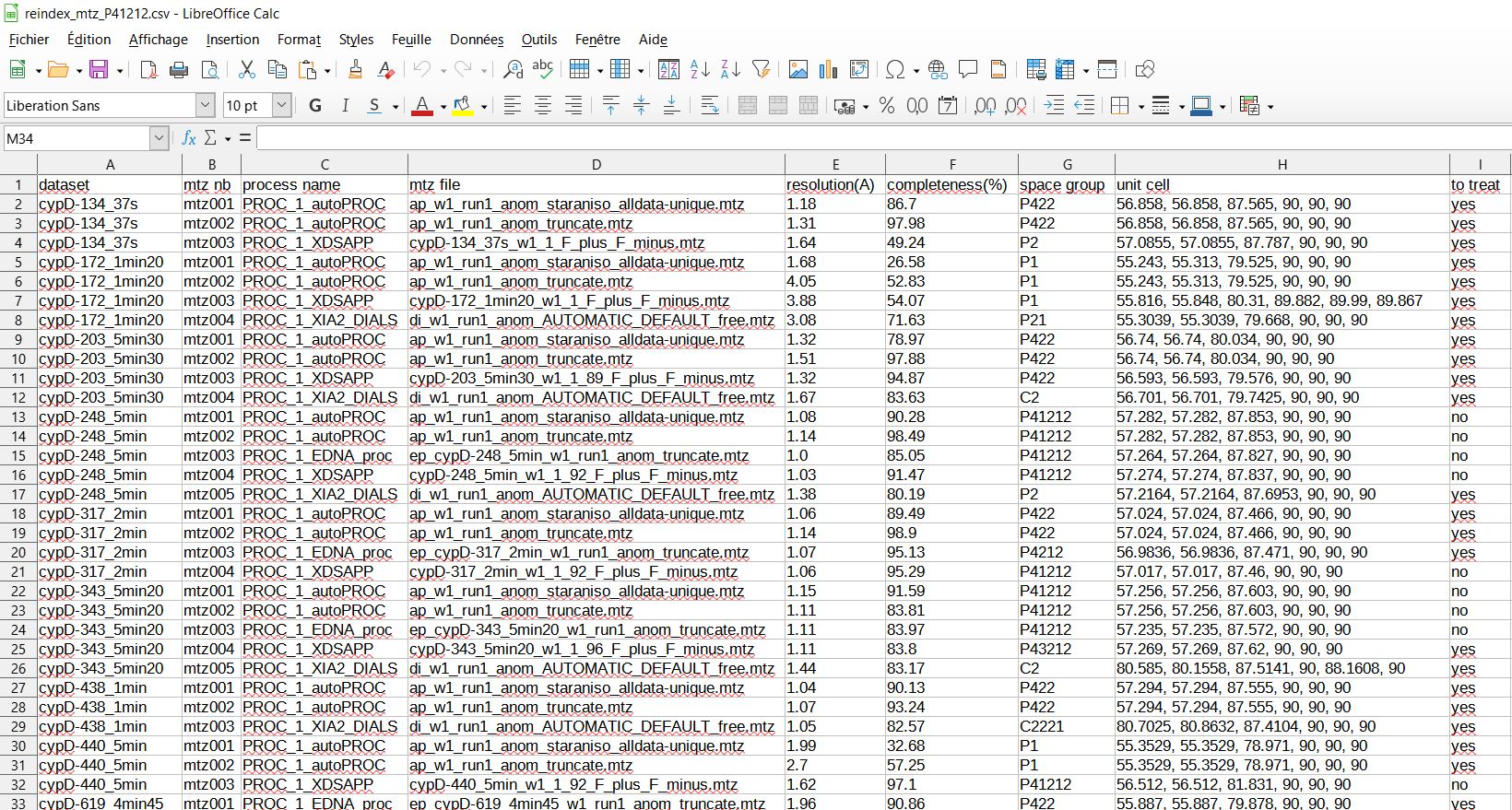
Example:
$ easypipe.py PROCESSED_DATA reindex P41212 --simulate
Warning
If you run again ‘prep’ step for any reason like adding new datasets, you will have to run again this ‘reindex’ step. Even if they don’t need to be reindexed, you have to run ‘reindex’ step to have the right reindexed csv file including these new datasets. For this, launch reindex again with simulate option to re-generate the csv file with new processes to treat, then launch again reindex.
