easYPipe ‘ligands’¶
This step is mandatory if you want Phenix to search ligand, else it is optional.
‘ligands’ subcommand generates pdb and cif from smiles with eLBOW [2] .
Important
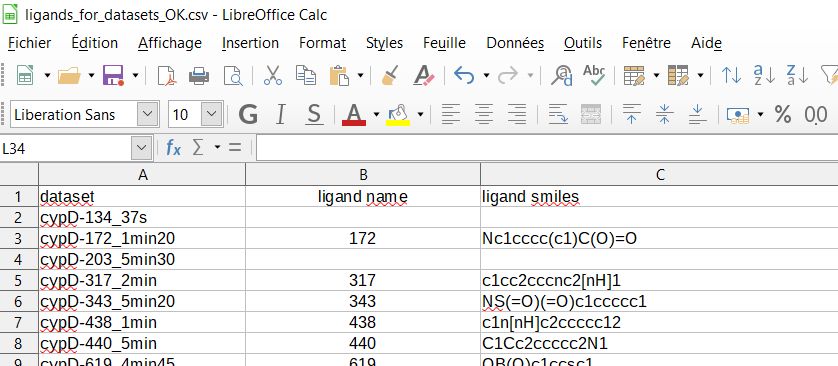
First, template csv file generated with ‘prep’ subcommand’ have to be completed with ligands names and smiles.
Usage¶
easypipe.py data ligands [-h] ligands_csv
arguments |
description |
|---|---|
-h, –help |
show this help message and exit |
ligands_csv |
ligands_for_datasets.csv file from ‘prep’ with ligands names and smiles completed (mandatory) |
Example:
$ easypipe.py PROCESSED_DATA ligands easYPipe/1c_ligands/ligands_for_datasets_OK.csv
What does it do ?¶
First, you have to fill in the fields ‘ligand name’ and ‘ligand smiles’ of /1c_ligands/ligands_for_datasets.csv csv file, then save the csv file somewhere else or with another name if you don’t want to overwrite it in case you run ‘prep’ subcommand again …
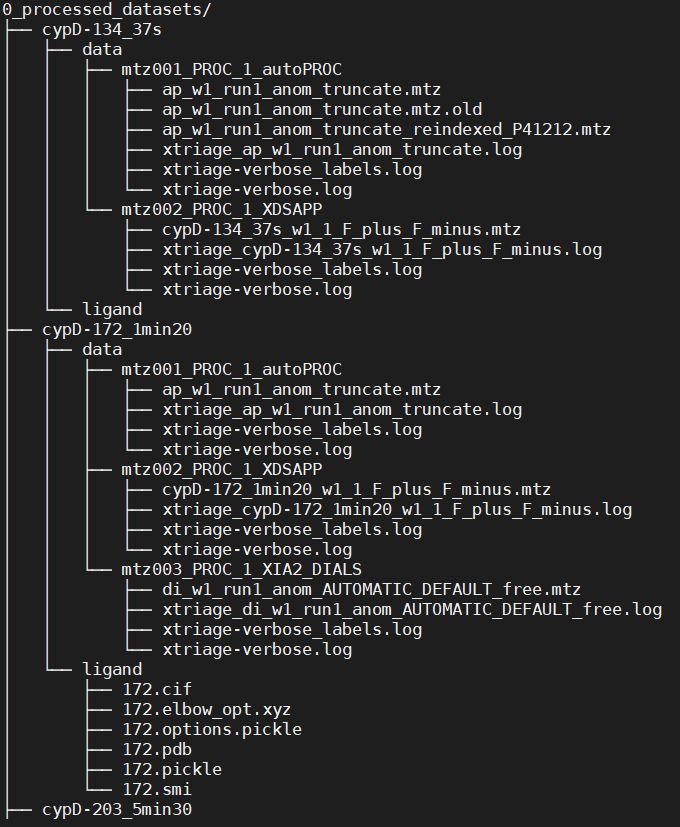
‘ligands’ subcommand generates pdb and cif of each ligand in a subfolder of the folder /1c_ligands/ligands and copies them in corresponding processed dataset folder (0_processed_datasets/’dataset name’), in a ‘ligand’ folder. It first creates a smiles file accordingly to the ligands_csv input, canonizes it thanks to Open Babel [1] and converts it with eLBOW [2] to pdb and cif.