easYPipe ‘summary’¶
This step can be run after several runs of ‘launch’ subcommands.
For each ‘launch’ subcommand, a ‘RESULT’ csv file is created that summarizes the corresponding results for each dataset (see here). So, if you have tried several options, you will have as many ‘RESULTS’ csv files.

Then, you probably want to compile all these results for a better view.
Now, the command ‘summary’ is automatically run at the end of each ‘launch’ command, but only for the RESULTS folder of this command (in this example, the ‘RESULTS_P41212’ folder).
If you have done several ‘launch’ with different space group for example, that means the RESULTS folders will be different, you will have to run manually the ‘summary’ command.
Then a global SUMMARY file will be created, that compiles all SUMMARY files present in RESULTS folders if there are several RESULTS folders (case when launched for several space groups, or different templates).
Usage¶
easypipe.py data summary [-h]
arguments |
description |
|---|---|
-h, –help |
show this help message and exit |
-o, –only_success |
generate also a result file with only successfull treatments |
Example:
$ easypipe.py PROCESSED_DATA summary
What does it do ?¶
In the ‘RESULT’ folder, ‘summary’ subcommand creates a ‘SUMMARY’ csv file where all datasets results are compiled.
With the option ‘–only_success’, unsucessfull treatments are not listed in the summary file, for a better clarity.
For each datasets, redondant results are deleted and the remaining ones are sorted according to:
‘dataset’
‘SUCCESS’
‘ligand search’
‘Ligand’ (found or not)
‘Completeness’
‘Nb of ligands found’
‘Rwork’
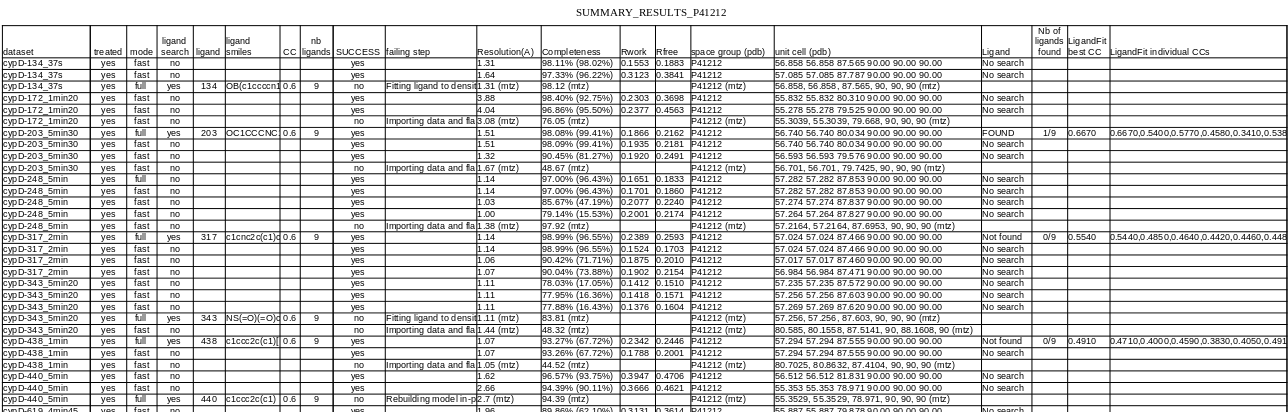
This way, the first row of each dataset should be most of the time the best treatment to consider, but is always better to have a critical eye on information like completeness or resolution to be sure …
And, if there are several RESULTS folders (case when launched for several space groups, or different templates), a global SUMMARY file will be created, that compiles all SUMMARY files present in RESULTS folders. Sorting criteria are the same as above.
