|
CCSP evaluation. |
|
Principle of use
CCSP submits a protonate PDB file (basic format) to the five following softwares: - Shifty v1.4 (Wishart et al, J Biomol NMR. 1997) - Orb v1.2 (Gronwald et al, J Biomol NMR. 1998) - Shifts v4.1.1 (Xu et al, J Biomol NMR. 2001) - Proshifts (Meiler, J Biomol NMR, 2003.) - Shiftx (Neal et al, J Biomol NMR. 2003) CCSP proposes a page containing the results of the softwares and a consensus result tab with mean, min and max values: Atom Shifty Orb Shifts ShiftX Proshifts Mean Min Max 1 A N ---- 123.80 ---- 123.35 122.83 123.33 122.83 123.35 1 A HN ---- 8.11 7.83 8.26 ---- 8.07 7.83 8.26 1 A HA1 4.01 4.19 3.68 4.09 4.22 4.04 3.68 4.22 1 A HB1 1.42 1.33 ---- 1.29 1.32 1.34 1.29 1.42 1 A HB2 ---- ---- 1.44 ---- 1.32 1.38 1.32 1.44 1 A CA ---- 52.47 ---- 51.53 51.61 51.87 51.53 51.61 1 A CB ---- 18.91 ---- 19.24 17.31 18.49 17.31 19.24 1 A CO ---- 177.12 ---- 176.28 176.16 176.52 176.16 176.28 2 C N 119.94 118.70 ---- 121.56 119.21 119.85 118.70 121.56 2 C HN 8.83 8.18 7.97 8.38 8.38 8.35 7.97 8.83 2 C HA1 4.65 4.52 4.13 3.89 4.73 4.38 3.89 4.73 2 C HB1 3.06 3.17 2.92 3.18 2.92 3.05 2.92 3.18 2 C HB2 2.73 2.80 2.92 3.15 2.88 2.90 2.73 3.15 2 C CA ---- 55.52 ---- 57.99 55.56 56.36 55.52 57.99 2 C CB ---- 42.00 ---- 36.19 33.04 37.08 33.04 36.19 2 C CO ---- 173.84 ---- 171.64 173.01 172.83 171.64 173.01 3 Q N 123.84 119.80 110.93 125.67 119.41 119.93 110.93 125.67 3 Q HN 7.85 8.19 8.04 9.06 8.25 8.28 7.85 9.06 3 Q HA1 4.58 4.28 4.46 3.94 4.29 4.31 3.94 4.58 3 Q HB1 2.00 2.12 1.94 2.05 1.98 2.02 1.94 2.05 3 Q HB2 1.81 1.95 1.81 2.15 1.98 1.94 1.81 2.15 3 Q CA ---- 55.62 55.84 56.87 54.09 55.61 54.09 56.87 3 Q CB ---- 30.16 28.23 28.88 26.86 28.53 26.86 28.88 3 Q CO ---- 176.06 178.72 176.97 174.46 176.55 174.46 178.72 4 A N 111.54 123.80 114.92 117.18 122.24 117.94 111.54 122.24 4 A HN 8.81 8.11 7.85 8.11 8.49 8.27 7.85 8.81 4 A HA1 3.62 4.19 3.76 3.77 4.25 3.92 3.62 4.25 4 A HB1 0.85 1.33 ---- 1.34 1.52 1.26 0.85 1.52 4 A HB2 ---- ---- 1.41 ---- 1.52 1.46 1.41 1.52 4 A CA ---- 52.47 53.52 52.38 51.11 52.37 51.11 53.52 4 A CB ---- 18.91 17.38 15.41 16.25 16.99 15.41 17.38 4 A CO ---- 177.12 177.93 177.67 174.48 176.80 174.48 177.93 |
Tests and evaluation
The tests were made from structures studied by the 2 techniques : Xray and NMR. Xray structures were used for the prediction and the NMR structures were used for the evaluation (BMRB file). The following tab lists the selected structures (1240 residues): |
| XRay PDB id (Date) |
NMR PDB id (Date) |
BMRB id |
Alpha Helix |
Beta Sheet |
| 3CBS (1999) |
1BM5 (1999) |
4186 |
12% |
54% |
| 1KMI (2002) |
1CEY (1995) |
4472 |
64% |
7% |
| 1IAR (2000) |
2CYK (1994) |
4094 |
30% |
31% |
| 1GZS (2002) |
1EES (2000) |
4700 |
53% |
14% |
| 1EL1 (2001) |
1I56 (2002) |
4876 |
36% |
8% |
| 1KQR (2002) |
1KRI (2002) |
5275 |
10% |
56% |
| 1MMS (2000) |
1OLN (2003) |
5513 |
42% |
15% |
| 1QJ8 (1999) |
1Q9F (2004) |
4936 |
no |
82% |
| 1BDO (1996) |
2BDO (1999) |
4425 |
no |
45% |
| 1MJC (1994) |
3MEF (1998) |
4296 |
4% |
49% |
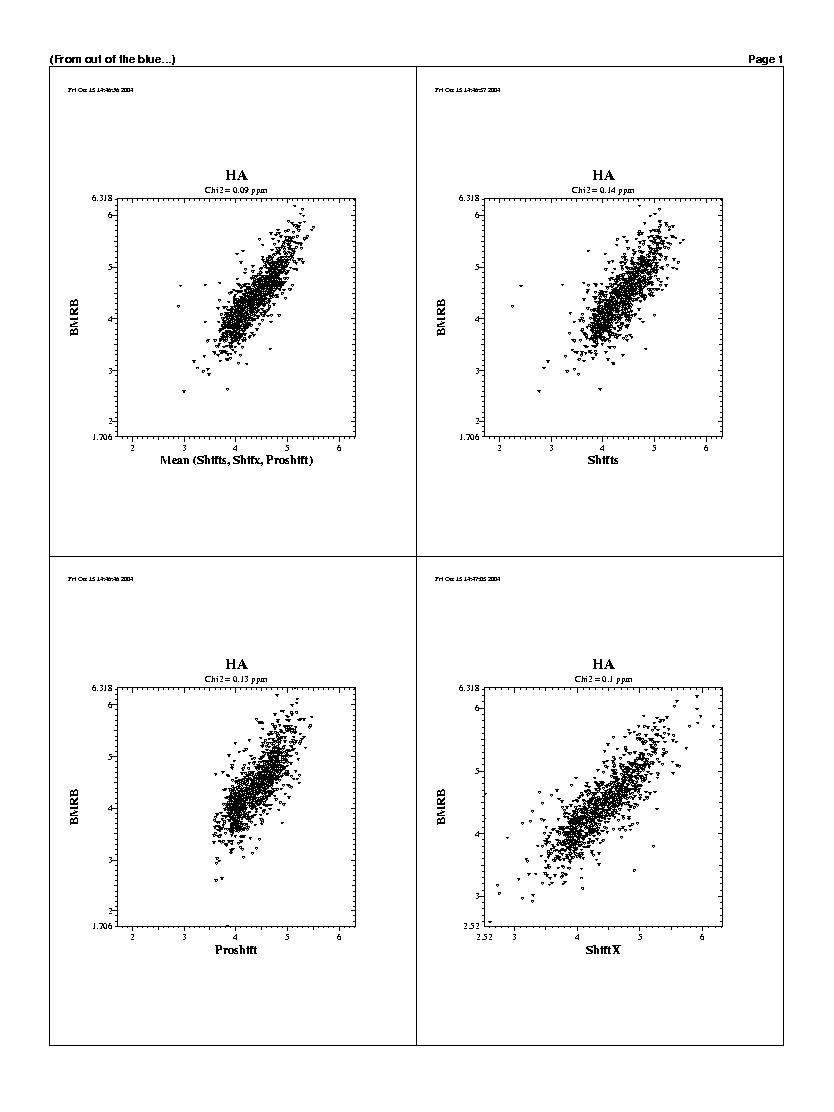
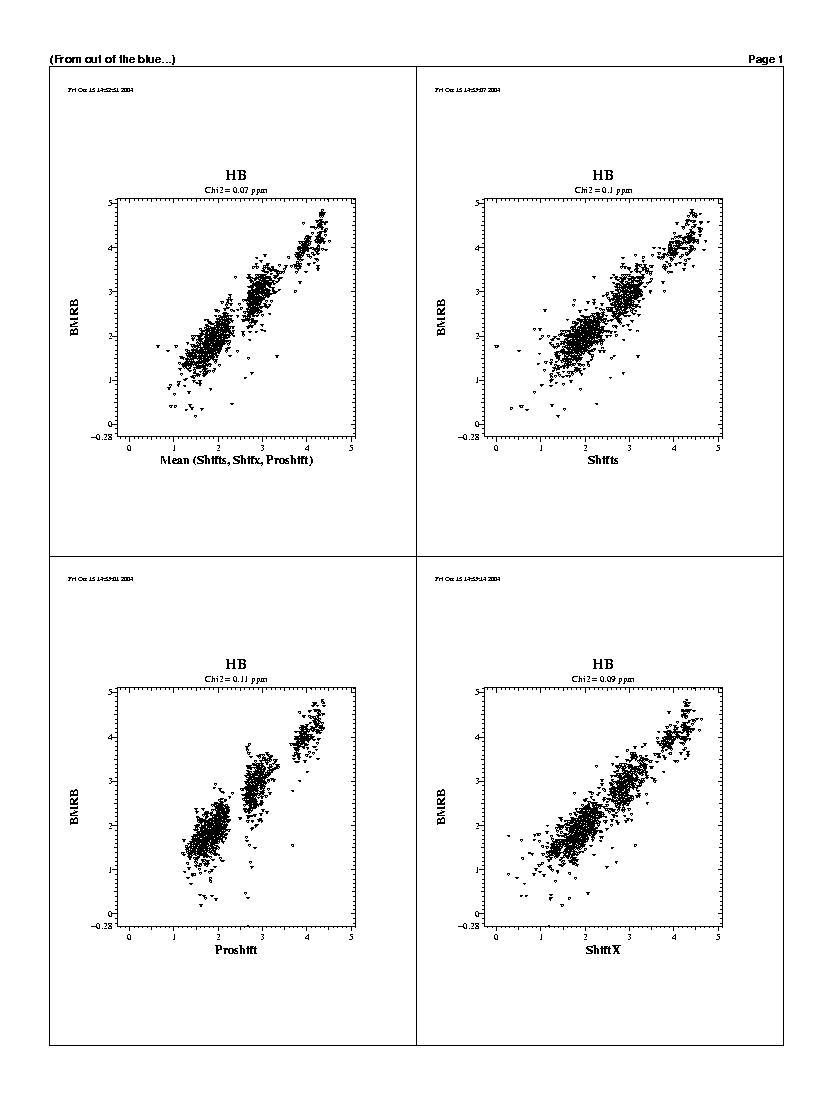
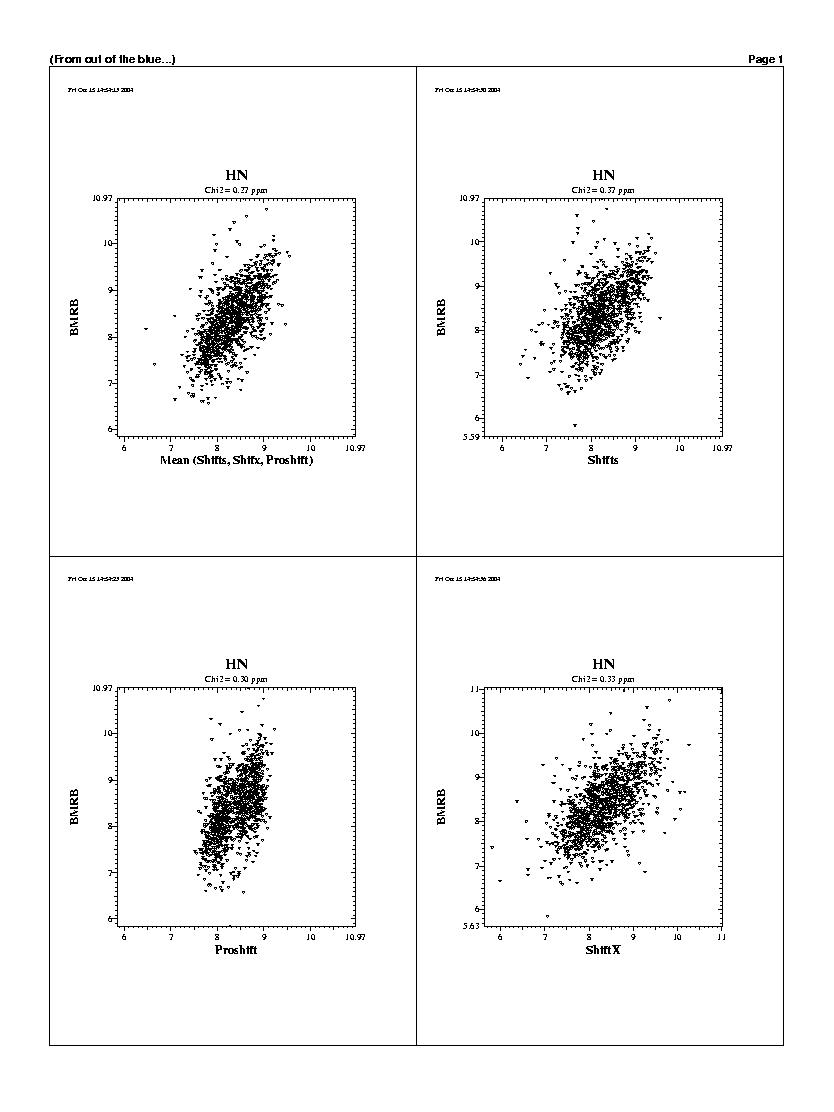
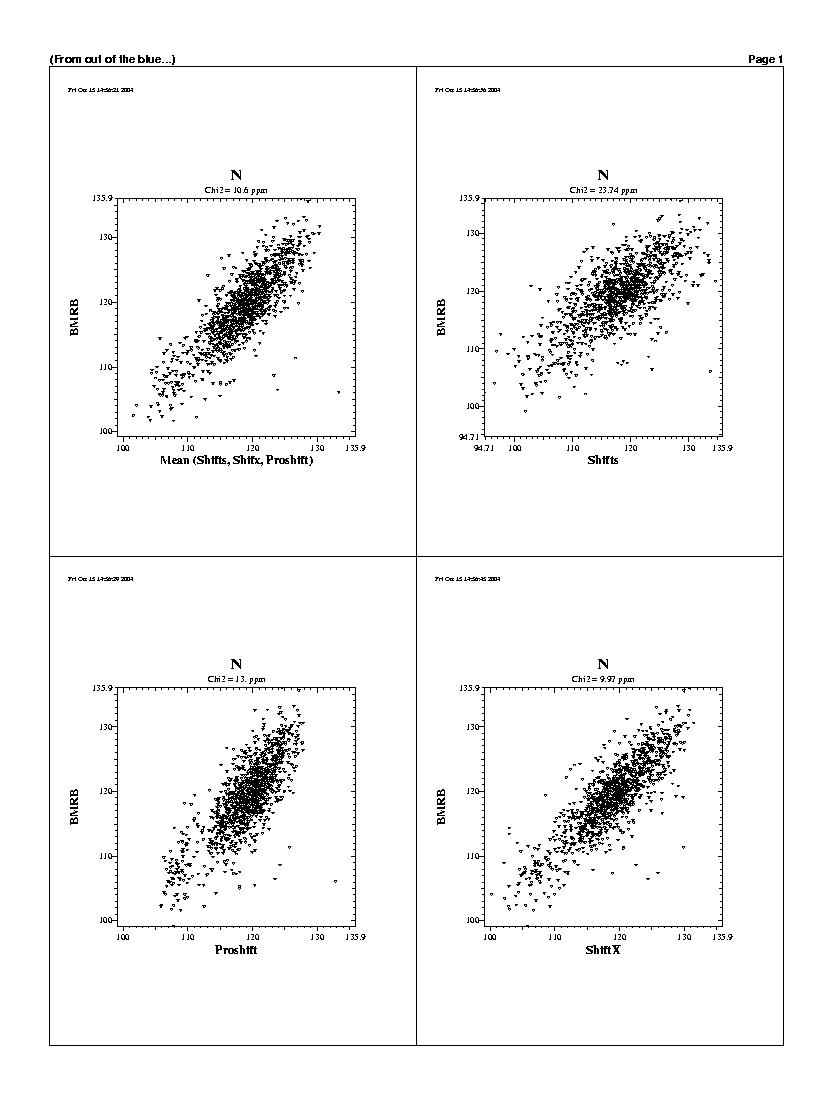
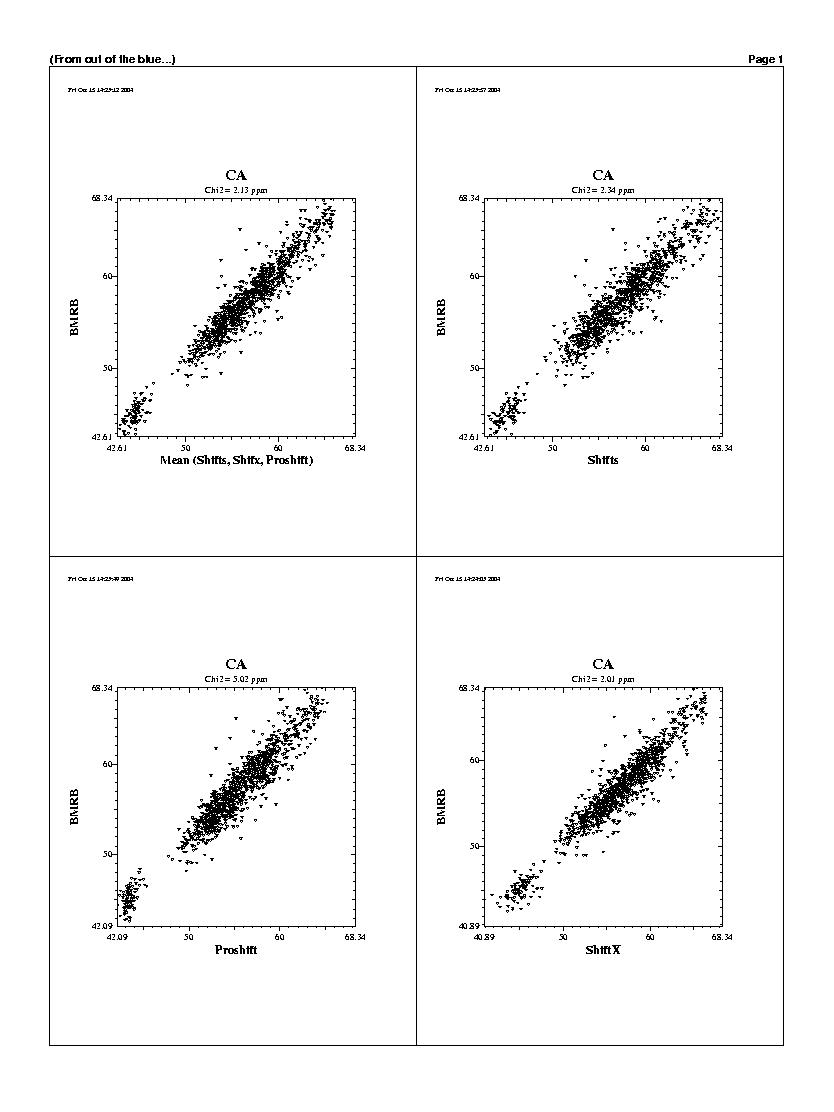
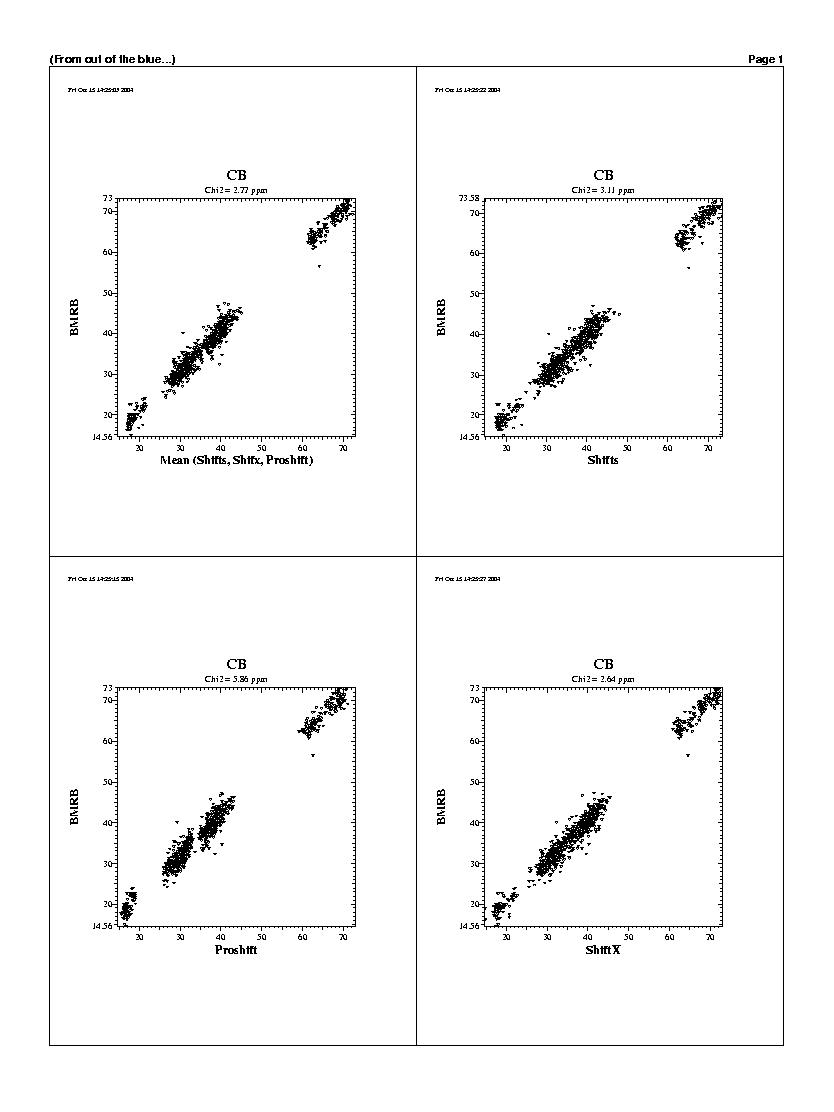
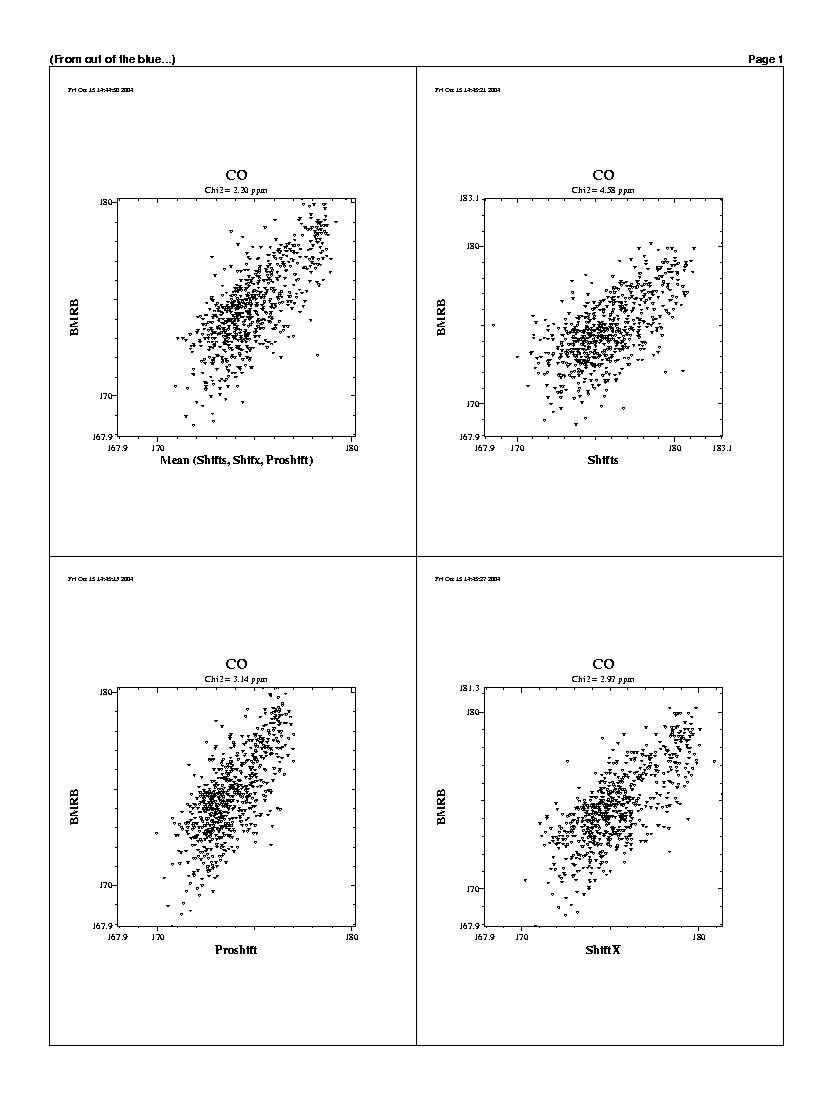
| The predicted chemicals shifts of the 1240 residues
are shown in the following graphs. For all backbones nucleis, Shifts, Proshifts,
ShiftX and CCSP results are available. The BMRB axe shows observed values.
Shifty and Orb results are not showed.
|






