Principle of use
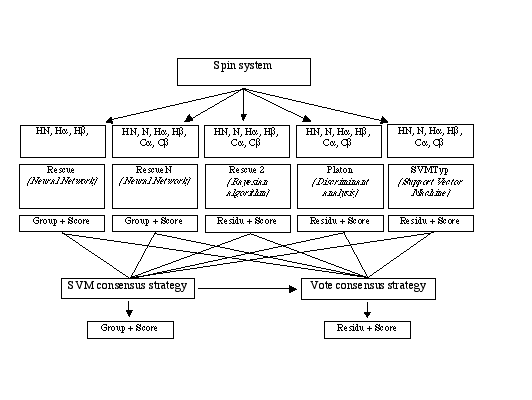
CRAACK submits a formated imput file to the five following typing softwares:
| Typing Tool |
Method |
Reference |
| Rescue 1 | Neural Network |
Pons et al (1999), J Biomol NMR. |
| Platon | Discriminant Analysis |
Labudde et al(2003). J Biomol NMR. |
| Rescue 1N | Neural Network | Auguin et al (2003), Spectroscopy. |
| Rescue 2 | Bayesian Algorithm |
Marin et al. (2003).J.Biomol.NMR. |
| SVMTyp | Support Vector Machine * |
JL Pons - not published |
* SVMTyp was built from SVMtorch (Ronan Collobert et al (2001) Journal of Machine Learning Research)
CRAACK is based on two consensus algorithm which tries to acccurate the responses of existant typing modules.
The first algorithm accomplishes the analysis with SVM technologie ( MySVM, Joachims, Thorsten (1999) Support Vector Learning, chapter 11. MIT Press).
The second one use a classical algorithm of vote.
Additionaly, secondary structural prediction is available (CSI :Wishart & Sykes. (1994) . J Biomol NMR)
Fig: Craack general flowchart:
CRAACK proposes a page containing the results of the softwares and a consensus result tab :
Num Modules Results
1 [Input Shifts]: (8.264; 115.78; 4.410; 4.263; 62.34; 70.01; 174.11; #1; T; )
[Rescue1]: G (67)
[Rescue1N]: T (39)
[Rescue2]: T (1.00)
[Platon]: T (56.22) S (44.55)
[SVMTyping]: T (5.33)
[SVM Consensus]: T (7.34)
[Vote Consensus]:T (5.4) G (1) S (0.4)
[CSI]: E(-1) E(-1) E(-1)